Getting Started with Conda
Overview
Teaching: 15 min
Exercises: 5 minQuestions
What is Conda?
Why should I use a package and environment management system as part of my research workflow?
Why use Conda ?
Objectives
Understand why you should use a package and environment management system as part of your (data) science workflow.
Explain the benefits of using Conda as part of your (data) science workflow.
Packages and Environments
Packages
When working with a programming language, such as Python, that can do almost anything, one has to wonder how this is possible. You download Python, it has about 25 MB, how can everything be included in this small data package. The answer is - it is not. Python, as well as many other programming languages use external libraries or packages for being able to doing almost anything. You can see this already when you start programming. After learning some very basics, you often learn how to import something into your script or session.
Modules, packages, libraries
- Module: a collection of functions and variables, as in a script
- Package: a collection of modules with an init.py file (can be empty), as in a directory with scripts
- Library: a collection of packages with related functionality
Library/Package are often used interchangeably.
Dependencies
A bit further into your programming career you may notice/have noticed that many packages do not just do everything on their own. Instead, they depend on other packages for their functionality. For example, the Scipy package is used for numerical routines. To not reinvent the wheel, the package makes use of other packages, such as numpy (numerical python) and matplotlib (plotting) and many more. So we say that numpy and matplotlib are dependencies of Scipy.
Many packages are being further developed all the time, generating different versions of packages. During development it may happen that a function call changes and/or functionalities are added or removed. If one package can depend on another, this may create issues. Therefore it is not only important to know that e.g. Scipy depends on numpy and matplotlib, but also that it depends on numpy version >= 1.6 and matplotlib version >= 1.1. Numpy version 1.5 in this case would not be sufficient.
Environments
When starting with programming we may not use many packages yet and the installation may be straightforward. But for most people, there comes a time when one version of a package or also the programming language is not enough anymore. You may find an older tool that depends on an older version of your programming language (e.g. Python 2.7), but many of your other tools depend on a newer version (e.g. Python 3.6). You could now start up another computer or virtual machine to run the other version of the programming language, but this is not very handy, since you may want to use the tools together in a workflow later on. Here, environments are one solution to the problem. Nowadays there are several environment management systems following a similar idea: Instead of having to use multiple computers or virtual machines to run different versions of the same package, you can install packages in isolated environments.
Environment management
An environment management system solves a number of problems commonly encountered by (data) scientists.
- An application you need for a research project requires different versions of your base programming language or different versions of various third-party packages from the versions that you are currently using.
- An application you developed as part of a previous research project that worked fine on your system six months ago now no longer works.
- Code that have written for a joint research project works on your machine but not on your collaborators’ machines.
- An application that you are developing on your local machine doesn’t provide the same results when run on your remote cluster.
An environment management system enables you to set up a new, project specific software environment containing specific Python versions as well as the versions of additional packages and required dependencies that are all mutually compatible.
- Environment management systems help resolve dependency issues by allowing you to use different versions of a package for different projects.
- Make your projects self-contained and reproducible by capturing all package dependencies in a single requirements file.
- Allow you to install packages on a host on which you do not have admin privileges.
Environment management systems for Python
Conda is not the only way; Python for example has many more ways of working with environments:
Package management
A good package management system greatly simplifies the process of installing software by…
- identifying and installing compatible versions of software and all required dependencies.
- handling the process of updating software as more recent versions become available.
If you use some flavor of Linux, then you are probably familiar with the package manager for your
Linux distribution (i.e., apt on Ubuntu, yum on CentOS); if you are a Mac OSX user then you
might be familiar with the Home Brew Project which brings a Linux-like package
management system to Mac OS; if you are a Windows OS user, then you may not be terribly familiar
with package managers as there isn’t really a standard package manager for Windows (although there
is the Chocolatey Project).
Operating system package management tools are great but these tools actually solve a more general problem than you often face as a (data) scientist. As a (data) scientist you typically use one or two core scripting languages (i.e., Python, R, SQL). Each scripting language has multiple versions that can potentially be installed and each scripting language will also have a large number of third-party packages that will need to be installed. The exact version of your core scripting language(s) and additional, third-party packages will also probably change from project to project.
Package management systems for Python
Also here, Conda is not the only way; Python for example has many more ways of working with packages:
Why should I use a package and environment management system?
Installing software is hard. Installing scientific software is often even more challenging. In order to minimize the burden of installing and updating software (data) scientists often install software packages that they need for their various projects system-wide.
Installing software system-wide has a number of drawbacks:
- It can be difficult to figure out what software is required for any particular research project.
- It is often impossible to install different versions of the same software package at the same time.
- Updating software required for one project can often “break” the software installed for another project.
Put differently, installing software system-wide creates complex dependencies between your research projects that shouldn’t really exist!
Rather than installing software system-wide, wouldn’t it be great if we could install software separately for each research project?
Discussion
What are some of the potential benefits from installing software separately for each project? What are some of the potential costs?
Solution
You may notice that many of the potential benefits from installing software separately for each project require the ability to isolate the projects’ software environments from one another (i.e., solve the environment management problem). Once you have figured out how to isolate project-specific software environments, you will still need to have some way to manage software packages appropriately (i.e., solve the package management problem).
What I hope you will have taken away from the discussion exercise is an appreciation for the fact that in order to install project-specific software environments you need to solve two complementary challenges: environment management and package management.
Conda
From the official Conda documentation. Conda is an open source package and environment management system that runs on Windows, Mac OS and Linux.
- Conda can quickly install, run, and update packages and their dependencies.
- Conda can create, save, load, and switch between project specific software environments on your local computer.
- Although Conda was created for Python programs, Conda can package and distribute software for any language such as R, Ruby, Lua, Scala, Java, JavaScript, C, C++, FORTRAN.
Conda as a package manager helps you find and install packages. If you need a package that requires a different version of Python, you do not need to switch to a different environment manager, because Conda is also an environment manager. With just a few commands, you can set up a totally separate environment to run that different version of Python, while continuing to run your usual version of Python in your normal environment.
Conda vs. Miniconda vs. Anaconda
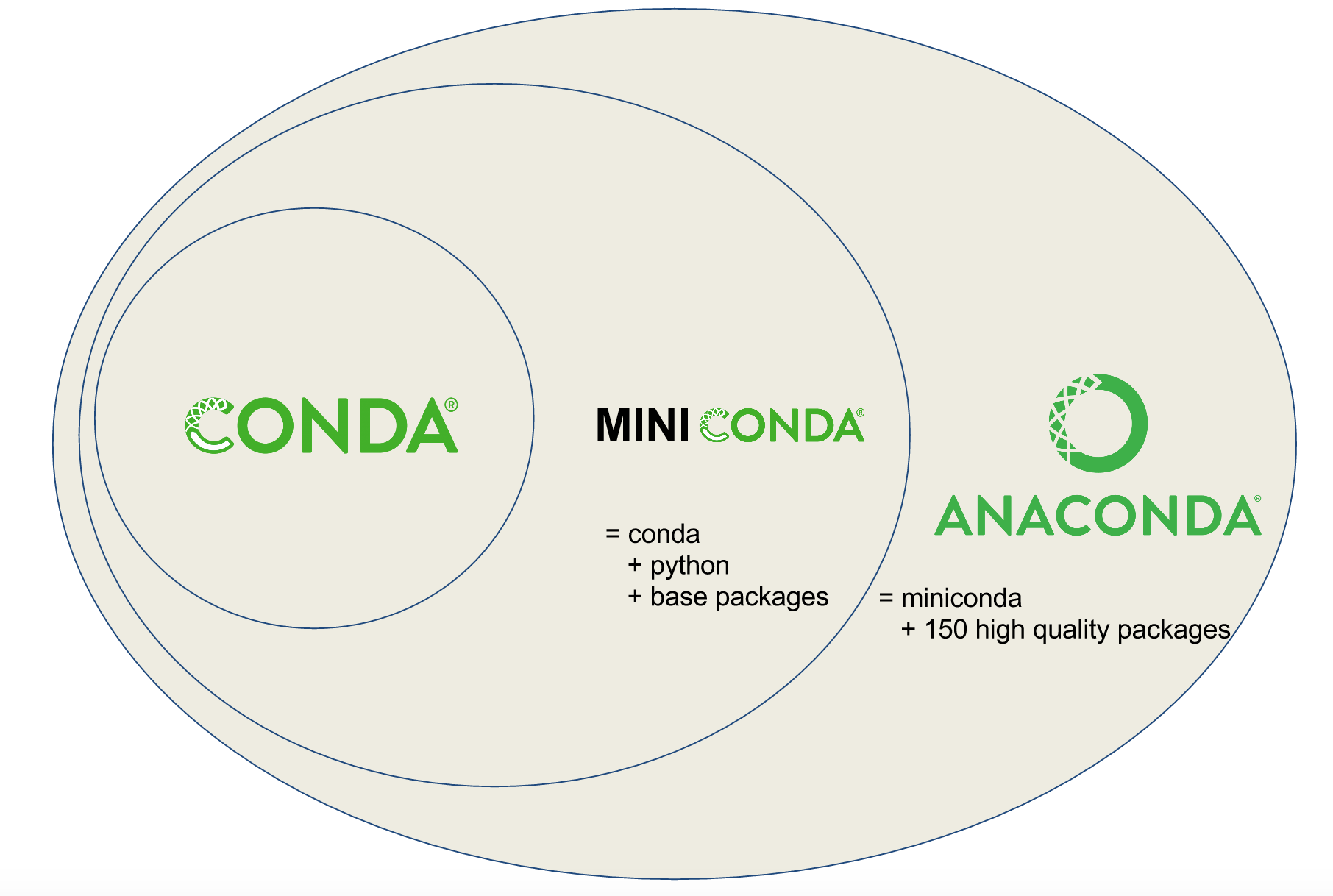
Users are often confused about the differences between Conda, Miniconda, and Anaconda. Conda is a tool for managing environments and installing packages. Miniconda combines Conda with Python and a small number of core packages; Anaconda includes Miniconda as well as a large number of the most widely used Python packages.
Why use Conda?
Whilst there are many different package and environment management systems that solve either the package management problem or the environment management problem, Conda solves both of these problems and explicitly targeted at (data) science use cases.
- Conda provides prebuilt packages, avoiding the need to deal with compilers, or trying to work out how exactly to set up a specific tool. Fields such as Astronomy use conda to distribute some of their most difficult-to-install tools such as IRAF. TensorFlow is another tool where to install it from source is near impossible, but Conda makes this a single step.
- Conda is cross platform, with support for Windows, MacOS, GNU/Linux, and support for multiple hardware platforms, such as x86 and Power 8 and 9. In future lessons we will show how to make your environment reproducible (reproducibility being one of the major issues facing science), and Conda allows you to provide your environment to other people across these different platforms.
- Conda allows for using other package management tools (such as
pip) inside Conda environments, where a library or tools is not already packaged for Conda (we’ll show later how to get access to more conda packages via channels).
Additionally, Anaconda provides commonly used data science libraries and tools, such as R, NumPy, SciPy and TensorFlow built using optimised, hardware specific libraries (such as Intel’s MKL or NVIDIA’s CUDA), which provides a speedup without having to change any of your code.
Key Points
Conda is a platform agnostic, open source package and environment management system.
Using a package and environment management tool facilitates portability and reproducibility of (data) science workflows.
Conda solves both the package and environment management problems and targets multiple programming languages. Other open source tools solve either one or the other, or target only a particular programming language.
Anaconda is not only for Python
Working with Environments
Overview
Teaching: 60 min
Exercises: 15 minQuestions
What is a Conda environment?
How do I create (delete) an environment?
How do I activate (deactivate) an environment?
How do I install packages into existing environments using Conda (+pip)?
Where should I create my environments?
How do I find out what packages have been installed in an environment?
How do I find out what environments that exist on my machine?
How do I delete an environment that I no longer need?
Objectives
Understand how Conda environments can improve your research workflow.
Create a new environment.
Activate (deactivate) a particular environment.
Install packages into existing environments using Conda (+pip).
Specify the installation location of an environment.
List all of the existing environments on your machine.
List all of the installed packages within a particular environment.
Delete an entire environment.
Workspace for Conda environments
If you haven’t done it yet, create a new
introduction-to-conda-for-data-scientistsdirectory on your Desktop in order to maintain a consistent workspace for all your conda environment.On Mac OSX and Linux running following commands in the Terminal will create the required directory on the Desktop.
$ cd ~/Desktop $ mkdir introduction-to-conda-for-data-scientists $ cd introduction-to-conda-for-data-scientistsFor Windows users you may need to reverse the direction of the slash and run the commands from the command prompt.
> cd ~\Desktop > mkdir introduction-to-conda-for-data-scientists > cd introduction-to-conda-for-data-scientistsAlternatively, you can always “right-click” and “create new folder” on your Desktop. All the commands that are run during the workshop should be run in a terminal within the
introduction-to-conda-for-data-scientistsdirectory.
What is a Conda environment
A Conda environment is a directory that contains a specific collection of Conda packages that you have installed. For example, you may be working on a research project that requires NumPy 1.18 and its dependencies, while another environment associated with an finished project has NumPy 1.12 (perhaps because version 1.12 was the most current version of NumPy at the time the project finished). If you change one environment, your other environments are not affected. You can easily activate or deactivate environments, which is how you switch between them.
Avoid installing packages into your
baseConda environmentConda has a default environment called
basethat include a Python installation and some core system libraries and dependencies of Conda. It is a “best practice” to avoid installing additional packages into yourbasesoftware environment. Additional packages needed for a new project should always be installed into a newly created Conda environment.
Creating environments
To create a new environment for Python development using conda you can use the conda create
command.
$ conda create --name python3-env python
For a list of all commands, take a look at Conda general commands.
It is a good idea to give your environment a meaningful name in order to help yourself remember
the purpose of the environment. While naming things can be difficult, $PROJECT-NAME-env is a
good convention to follow. Sometimes also the specific version of a package why you had to create a new environment is a good name
The command above will create a new Conda environment called “python3-env” and install the most recent version of Python. If you wish, you can specify a particular version of packages for conda to install when creating the environment.
$ conda create --name python36-env python=3.6
Always specify a version number for each package you wish to install
In order to make your results more reproducible and to make it easier for research colleagues to recreate your Conda environments on their machines it is a “best practice” to always explicitly specify the version number for each package that you install into an environment. If you are not sure exactly which version of a package you want to use, then you can use search to see what versions are available using the
conda searchcommand.$ conda search $PACKAGE_NAMESo, for example, if you wanted to see which versions of Scikit-learn, a popular Python library for machine learning, were available, you would run the following.
$ conda search scikit-learnAs always you can run
conda search --helpto learn about available options.
You can create a Conda environment and install multiple packages by listing the packages that you wish to install.
$ conda create --name basic-scipy-env ipython=7.13 matplotlib=3.1 numpy=1.18 scipy=1.4
When conda installs a package into an environment it also installs any required dependencies.
For example, even though Python is not listed as a packaged to install into the
basic-scipy-env environment above, conda will still install Python into the environment
because it is a required dependency of at least one of the listed packages.
Creating a new environment
Create a new environment called “machine-learning-env” with Python and the most current versions of IPython, Matplotlib, Pandas, Numba and Scikit-Learn.
Solution
In order to create a new environment you use the
conda createcommand as follows.$ conda create --name machine-learning-env \ ipython \ matplotlib \ pandas \ python \ scikit-learn \ numbaSince no version numbers are provided for any of the Python packages, Conda will download the most current, mutually compatible versions of the requested packages. However, since it is best practice to always provide explicit version numbers, you should prefer the following solution.
$ conda create --name machine-learning-env \ ipython=7.19 \ matplotlib=3.3 \ pandas=1.2 \ python=3.8 \ scikit-learn=0.23 \ numba=0.51However, please be aware that the version numbers for each packages may not be the latest available and would need to be adjusted.
Renaming a conda environment
As of conda version 4.14.0 you can rename a conda environment with the
conda renamecommand. conda rename supports renaming your current environment, or any of your existing environments.conda rename -n old_env_name new_env_name
Activating an existing environment
Activating environments is essential to making the software in environments work well (or sometimes at all!). Activation of an environment does two things.
- Adds entries to
PATHfor the environment. - Runs any activation scripts that the environment may contain.
Step 2 is particularly important as activation scripts are how packages can set arbitrary
environment variables that may be necessary for their operation. You activate the
basic-scipy-env environment by name using the activate command.
$ conda activate basic-scipy-env
You can see that an environment has been activated because the shell prompt will now include the name of the active environment.
(basic-scipy-env) $
Deactivate the current environment
To deactivate the currently active environment use the Conda deactivate command as follows.
(basic-scipy-env) $ conda deactivate
You can see that an environment has been deactivated because the shell prompt will no longer include the name of the previously active environment.
Returning to the
baseenvironmentTo return to the
baseConda environment, it’s better to callconda activatewith no environment specified, rather than to usedeactivate. If you runconda deactivatefrom yourbaseenvironment, you may lose the ability to runcondacommands at all. Don’t worry if you encounter this undesirable state! Just start a new shell.
Activate an existing environment by name
Activate the
machine-learning-envenvironment created in the previous challenge by name.Solution
In order to activate an existing environment by name you use the
conda activatecommand as follows.$ conda activate machine-learning-env
Deactivate the active environment
Deactivate the
machine-learning-envenvironment that you activated in the previous challenge.Solution
In order to deactivate the active environment you use the
conda deactivatecommand.(active-environment-name) $ conda deactivate
Installing a package into an existing environment
You can install a package into an existing environment using the conda install command. This command accepts a list of package specifications (i.e., numpy=1.18) and installs a set of packages consistent with those specifications and compatible with the underlying environment. If full compatibility cannot be assured, an error is reported and the environment is not changed.
By default the conda install command will install packages into the current, active environment.
The following would activate the basic-scipy-env we created above and install
Numba, an open source JIT compiler that translates a subset of Python
and NumPy code into fast machine code, into the active environment.
$ conda activate basic-scipy-env
$ conda install numba
As was the case when listing packages to install when using the conda create command, if version
numbers are not explicitly provided, Conda will attempt to install the newest versions of any
requested packages. To accomplish this, Conda may need to update some packages that are already
installed or install additional packages. It is always a good idea to explicitly provide version
numbers when installing packages with the conda install command. For example, the following would
install a particular version of Scikit-Learn, into the current, active environment.
$ conda install scikit-learn=0.22
Freezing installed packages
To prevent existing packages from being updating when using the
conda installcommand, you can use the--freeze-installedoption. This may force Conda to install older versions of the requested packages in order to maintain compatibility with previously installed packages. Using the--freeze-installedoption does not prevent additional dependency packages from being installed.
Remove a package from an environment
To remove a package from an environment you can run the command.
$ conda uninstall PKGNAME --name ENVNAME
For example to remove the scikit-learn package from the basic-scipy-env environment run
$ conda uninstall scikit-learn --name basic-scipy-env
Installing a package into a specific environment
Dask provides advanced parallelism for data science workflows enabling performance at scale for the core Python data science tools such as Numpy Pandas, and Scikit-Learn. Have a read through the official documentation for the
conda installcommand and see if you can figure out how to install Dask into themachine-learning-envthat you created in the previous challenge.Solution
You can install Dask into
machine-learning-envusing theconda installcommand as follow.$ conda install --name machine-learning-env dask=2020.12You could also install Dask into
machine-learning-envby first activating that environment and then using theconda installcommand.$ conda activate machine-learning-env $ conda install dask=2020.12
Where do Conda environments live?
Environments created with conda, by default, live in the envs/ folder of your miniconda3 (or anaconda3) directory the absolute path to which will look something the following: /Users/$USERNAME/miniconda3/envs or C:\Users\$USERNAME\Anaconda3.
You can see the location of your conda environments by running the command.
conda config --show envs_dirs
Running ls (linux) / dir (Windows) on your anaconda envs/ directory will list out the directories containing the existing Conda environments.
Location of Conda environments on Binder
If you are working through these lessons using a Binder instance, then the default location of the Conda environments is slightly different.
$ /srv/conda/envsRunning
ls /srv/conda/envs/from a terminal will list out the directories containing any previously installed Conda environments.
How do I specify a location for a Conda environment?
You can control where a Conda environment lives by providing a path to a target directory when
creating the environment. For example the following command will create a new environment in a
sub-directory of the current working directory called env.
$ conda create --prefix ./env ipython=7.13 matplotlib=3.1 pandas=1.0 python=3.6
dot-slash, ./
In unix the dot-slash, ./, is a relative path a file or directory in the current directory.
You activate an environment created with a prefix using the same command used to activate environments created by name.
$ conda activate ./env
It is often a good idea to specify a path to a sub-directory of your project directory when creating an environment. Why?
- Makes it easy to tell if your project utilizes an isolated environment by including the environment as a sub-directory.
- Makes your project more self-contained as everything including the required software is contained in a single project directory.
An additional benefit of creating your project’s environment inside a sub-directory is that you
can then use the same name for all your environments; if you keep all of your environments in
your ~/miniconda3/env/ folder, you’ll have to give each of them a different name.
Conda environment sub-directory naming convention
In order to be consistent with the convention used by tools such as
venvandPipenv, I recommend usingenvas the name of the sub-directory of your project directory that contains your Conda environment. A benefit of maintaining the convention is that your environment sub-directory will be automatically ignored by the default Python.gitignorefile used on GitHub.Whatever naming convention you adopt it is important to be consistent! Using the same name for all of your Conda environments allows you to use the same
activatecommand as well.$ cd my-project/ $ conda activate ./env
Creating a new environment as a sub-directory within a project directory
First create a project directory called
project-dirusing the following command.$ mkdir project-dir $ cd project-dirNext, create a new environment inside the newly created
project-dirin a sub-directory calledenvan install Python 3.6, version 3.1 of Matplotlib, and version 2.0 of TensorFlow.Solution
project-dir $ conda create --prefix ./env \ python=3.6 \ matplotlib=3.1 \ tensorflow=2.0
Placing Conda environments outside of the default ~/miniconda3/envs/ folder comes with a couple of minor drawbacks. First, conda can no longer find your environment with the --name flag; you’ll generally need to pass the --prefix flag along with the environment’s full path to find the environment.
Second, an annoying side-effect of specifying an install path when creating your Conda environments is that your command prompt is now prefixed with the active environment’s absolute path rather than the environment’s name. After activating an environment using its prefix your prompt will look similar to the following.
(/absolute/path/to/env) $
As you can imagine, this can quickly get out of hand.
(/Users/USER_NAME/research/data-science/PROJECT_NAME/env) $
If (like me!) you find this long prefix to your shell prompt annoying, then there is a quick fix:
modify the env_prompt setting in your .condarc file, which you can do with the following
command.
$ conda config --set env_prompt '({name})'
This will either edit your ~/.condarc file if you already have one or create a ~/.condarc file
if you do not. Now your command prompt will display the active environment’s generic name.
$ cd project-directory
$ conda activate ./env
(env) project-directory $
For more on modifying your .condarc file, see the
official Conda docs.
Activate an existing environment by path
Activate the environment created in a previous challenge using the path to the environment directory.
Solution
You can activate an existing environment by providing the path the the environment directory instead of the environment name when using the
conda activatecommand as follows.$ conda activate ./envNote that the provided path can either be absolute or relative. If the path is a relative path then it must start with
./on Unix systems and.\when using PowerShell on Windows.
Conda can create environments for R projects too!
First create a project directory called
r-project-dirusing the following command.cd ~/ mkdir r-project-dir cd r-project-dirNext, take a look through the list of R packages available by default for installation using
conda. Create a new environment inside the newly createdr-project-dirin a sub-directory calledenvand installr-base,r-tidyverse.Solution
conda create --prefix ./env \ r-base \ r-tidyverse
Listing existing environments
Now that you have created a number of Conda environments on your local machine you have probably
forgotten the names of all of the environments and exactly where they live. Fortunately, there is
a conda command to list all of your existing environments together with their locations.
$ conda env list
Listing the contents of an environment
In addition to forgetting names and locations of Conda environments, at some point you will
probably forget exactly what has been installed in a particular Conda environment. Again, there is
a conda command for listing the contents on an environment. To list the contents of the
basic-scipy-env that you created above, run the following command.
$ conda list --name basic-scipy-env
If you created your Conda environment using the --prefix option to install packages into a
particular directory, then you will need to use that prefix in order for conda to locate the
environment on your machine.
$ conda list --prefix /path/to/conda-env
Listing the contents of a particular environment.
List the packages installed in the
machine-learning-envenvironment that you created in a previous challenge.Solution
You can list the packages and their versions installed in
machine-learning-envusing theconda listcommand as follows.$ conda list --name machine-learning-envTo list the packages and their versions installed in the active environment leave off the
--nameor--prefixoption.$ conda list
Deleting entire environments
Occasionally, you will want to delete an entire environment. Perhaps you were experimenting with
conda commands and you created an environment you have no intention of using; perhaps you no
longer need an existing environment and just want to get rid of cruft on your machine. Whatever
the reason, the command to delete an environment is the following.
$ conda remove --name python36-env --all
If you wish to delete and environment that you created with a --prefix option, then you will
need to provide the prefix again when removing the environment.
$ conda remove --prefix /path/to/conda-env/ --all
Delete an entire environment
Delete the entire “machine-learning-env” environment.
Solution
In order to delete an entire environment you use the
conda removecommand as follows.$ conda remove --name machine-learning-env --all --yesThis command will remove all packages from the named environment before removing the environment itself. The use of the
--yesflag short-circuits the confirmation prompt (and should be used with caution).
Key Points
A Conda environment is a directory that contains a specific collection of Conda packages that you have installed.
You create (remove) a new environment using the
conda create(conda remove) commands.You activate (deactivate) an environment using the
conda activate(conda deactivate) commands.You install packages into environments using
conda install; you install packages into an active environment usingpip install.You should install each environment as a sub-directory inside its corresponding project directory
Use the
conda env listcommand to list existing environments and their respective locations.Use the
conda listcommand to list all of the packages installed in an environment.
Using Packages and Channels
Overview
Teaching: 20 min
Exercises: 10 minQuestions
What are Conda packages?
What are Conda channels?
Why should I be explicit about which channels my research project uses?
What should I do if a Python package isn’t available via a Conda channel?
Objectives
Install a package from a specific channel.
Understand how conda channels work.
Use pip to install a package into your environment.
What are Conda packages?
A conda package is a compressed archive file (.tar.bz2) that contains:
- system-level libraries
- Python or other modules
- executable programs and other components
- metadata under the
info/directory - a collection of files that are installed directly into an
installprefix.
Conda keeps track of the dependencies between packages and platforms; the conda package format is identical across platforms and operating systems.
Package Structure
All conda packages have a specific sub-directory structure inside the tarball file. There is a
bin directory that contains any binaries for the package; a lib directory containing the
relevant library files (i.e., the .py files); and an info directory containing package metadata.
For a more details of the conda package specification, including discussions of the various
metadata files, see the [docs][conda-pkg-spec-docs].
As an example of Conda package structure consider the Conda package for
the RNA-Seq transcript quantification package salmon targeting a 64-bit Linux, salmon-1.4.0-h84f40af_1.tar.bz2.
.
├── bin
│ └── salmon
├── info
│ ├── about.json
│ ├── files
│ ├── git
│ ├── hash_input.json
│ ├── has_prefix
│ ├── index.json
│ ├── licenses
│ │ └── LICENSE
│ ├── paths.json
│ ├── recipe
│ │ ├── 0.14.2-1
│ │ │ ├── build.sh
│ │ │ └── meta.yaml
│ │ ├── build.sh
│ │ ├── conda_build_config.yaml
│ │ ├── meta.yaml
│ │ ├── meta.yaml.template
│ │ └── run_test.sh
│ ├── repodata_record.json
│ └── test
│ ├── run_test.sh
│ └── sample_data.tgz
└── lib
├── graphdump
│ ├── graphdump-targets.cmake
│ └── graphdump-targets-release.cmake
├── libgraphdump.a
├── libntcard.a
├── libsalmon_core.a
├── libtwopaco.a
├── ntcard
│ ├── ntcard-targets.cmake
│ └── ntcard-targets-release.cmake
└── twopaco
├── twopaco-targets.cmake
└── twopaco-targets-release.cmake
Conda packages cache directory (pkgs_dirs)
When you first install a package, conda will download the tar.bz2 packages into the package cache directory. Before conda install a package it will look in the package directory . By default this will be placed under ~/.conda/pkgs.
For Anaconda 5.0.1 and newer, you can also configure your pkgs directory location by using the following command:conda config --add pkgs_dirs <package directory>
What actually happens when I install packages?
During the installation process, files are extracted into the specified environment (defaulting to the current environment if none is specified). Installing the files of a conda package into an environment can be thought of as changing the directory to an environment, and then downloading and extracting the package and its dependencies.
For example, when you conda install a package that exists in a known repository (channel), and has no dependencies,
conda does the following.
- looks at your configured channels (in priority)
- reaches out to the repodata associated with your channels/platform
- parses repodata to search for the package
- once the package is found, conda pulls it down and installs
The conda documentation has a nice decision tree that describes the package installation process.
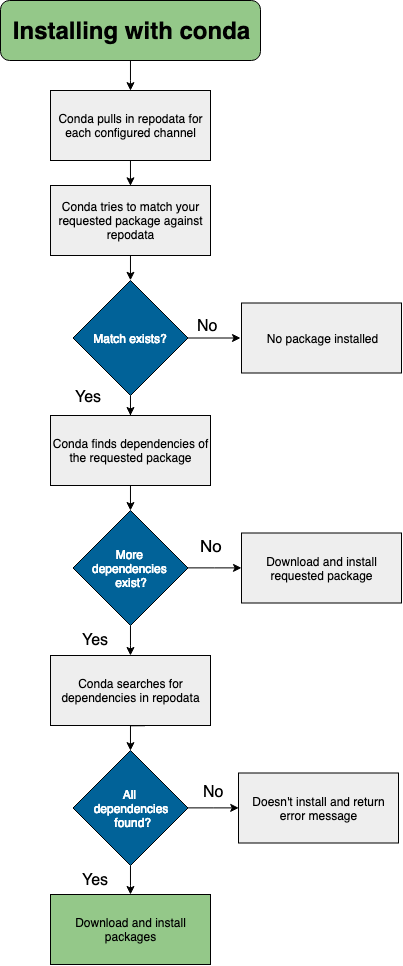
Channel
Let’s create a new environment basic-rnaseq-env and install a transcript quantification package, salmon , for an RNA-Seq analysis project.
$conda create --name basic-rnaseq-env salmon
This will return;
Loading channels: done
No match found for: salmon. Search: *salmon*
PackagesNotFoundError: The following packages are not available from current channels:
- salmon
Current channels:
- https://repo.anaconda.com/pkgs/main/linux-64
- https://repo.anaconda.com/pkgs/main/noarch
- https://repo.anaconda.com/pkgs/r/linux-64
- https://repo.anaconda.com/pkgs/r/noarch
To search for alternate channels that may provide the conda package you're
looking for, navigate to
https://anaconda.org
and use the search bar at the top of the page.
What does packages are not available from current channels: mean?
What are Conda channels?
When you install or search for a package in conda it searches for it in remote repositories called channels. These remote channel are URLs to directories containing conda packages. By default the conda search command searches a set of channels defined here.
Anaconda Cloud channels.
main: The majority of all new Anaconda, Inc. package builds are hosted here. Included in conda’s defaults channel as the top priority channel.r: Microsoft R Open conda packages and Anaconda, Inc.’s R conda packages. This channel is included in conda’s defaults channel. When creating new environments, MRO is now chosen as the default R implementation.
Collectively, the Anaconda managed channels are referred to as the defaults channel because, unless otherwise specified, packages installed using conda will be downloaded from these channels.
My package isn’t available on the defaults channel! What should I do?
As was the case with salmon it may very well be the case that packages (or often more recent versions of packages!) that you need to install for your project are not available on the defaults channel. In this case you should search for alternate channels that may provide the conda package you’re looking for, To do this you should first navigate to
https://anaconda.org
and use the search bar at the top of the page. If we search for salmon we will se it is available via a channel called bioconda.
bioconda
Bioconda is a channel, maintained by the Bioconda project, specialising in bioinformatics software. Bioconda contains 1000’s of bioinformatics packages ready to use with conda install.
R and Bioconductor packages Most R packages on CRAN should be submitted at Conda-Forge. However, if the CRAN package has a Bioconductor, a repository for bioinformatics R packages, dependency, it belongs in Bioconda. If the CRAN package does not have a Bioconductor package dependency, it belongs in Conda-Forge.
conda-forge
In addition to the default channels that are managed by Anaconda Inc., there is another channel called Conda-Forge that also has a special status. The Conda-Forge project “is a community led collection of recipes, build infrastructure and distributions for the conda package manager.”
There are a number of reasons that you may wish to use the conda-forge channel instead of the defaults channel maintained by Anaconda:
- Packages on
conda-forgemay be more up-to-date than those on thedefaultschannel. - There are packages on the
conda-forgechannel that aren’t available fromdefaults. - You may wish to use a dependency such as
openblas(fromconda-forge) instead ofmkl(fromdefaults).
How do I search for a package from a specific channel?
If you know the channel your package is likely to be located on, you can can use the conda search
command with the --channel option and the name of the channel. E.g.
$ conda search --channel bioconda salmon
Loading channels: done
# Name Version Build Channel
salmon 0.5.1 0 bioconda
salmon 0.6.0 0 bioconda
...[truncated]...
salmon 1.3.0 hf69c8f4_0 bioconda
salmon 1.4.0 h84f40af_1 bioconda
salmon 1.4.0 hf69c8f4_0 bioconda
salmon 1.5.0 h84f40af_0 bioconda
How do I install a package from a specific channel?
If you know the channel your package is available from you can install a package from a specific channel into the currently activate environment by passing the --channel or -c option to the conda install command as follows.
$ conda create --name basic-rnaseq-env
$ conda activate basic-rnaseq-env
$ conda install --channel bioconda salmon
You can also install a package from a specific channel into a named environment (using --name or -n) or into an environment installed at a particular prefix (using --prefix or -p). For example, the following command installs the salmon package from the bioconda channel into the environment called basic-rnaseq-env which we created earlier.
$ conda install salmon --channel bioconda --name basic-rnaseq-env
This command would install salmon package from bioconda channel into an environment
installed into the env/ sub-directory.
$ conda install salmon --channel bioconda --prefix ./env
You may have noticed that we didn’t manage to install the latest version fo salmon, why?
The bioconda channel contains bioinformatics packages (salmon, STAR, samtools, DESeq2, etc.), however the channel conda-forge has most of the dependencies (numpy, scipy, zlib, CRAN packages, etc.) needed. Therefore we need to specify multiple channels to install the latest version.
Specify multiple channels
To specify multiple channels for installing packages by passing the --channel argument
multiple times.
$ conda install salmon=1.5 --channel conda-forge --channel bioconda --name basic-rnaseq-env
This also works when install multiple packages.
$ conda install fastqc=0.11 multiqc=1.10 --channel conda-forge --channel bioconda --name basic-rnaseq-env
Specifying channels when installing packages
Create a new directory called
rnaseq-projectand then create an environment in a sub-directory calledenv/with the the packages salmon=1.5, fastqc=0.11 and multiqc=1.11.Solution
In order to create a new environment you use the
conda createcommand as follows.cd ~/ mkdir rnaseq-project cd rnaseq-project/ conda create --prefix ./env --channel conda-forge \ --channel bioconda \ salmon=1.5 \ fastqc=0.11 \ multiqc=1.11Hint: For the lazy typers: the
--channelargument can also be shortened to-c, for more abbreviations, see also the Conda command reference .
Alternative syntax for installing packages from specific channels
There exists an alternative syntax for installing conda packages from specific channels that more explicitly links the channel being used to install a particular package.
$ conda install biconda::multiqc --prefix ./envInstall the latest version of the workflow manager nextflow using this alternative syntax
Solution
One possibility would be to use the
conda createcommand as follows.$ conda install --prefix ./env --channel conda-forge bioconda::nextflow
Channel priority
Different channels can have the same package, so conda must decide which channel to install the package from. Conda channels have a priority hierarchy.
By default, conda prefers packages from a higher priority channel over any version from a lower priority channel.
Conda collects all of the packages with the same name across all listed channels and processes them as follows:
-
Sorts packages from highest to lowest channel priority.
-
Sorts tied packages—packages with the same channel priority—from highest to lowest version number. For example, if channelA contains NumPy 1.12.0 and 1.13.1, NumPy 1.13.1 will be sorted higher.
-
Sorts still-tied packages, packages with the same channel priority and same version, from highest to lowest build number. For example, if channelA contains both NumPy 1.12.0 build 1 and build 2, build 2 is sorted first. Any packages in channelB would be sorted below those in channelA.
-
Installs the first package on the sorted list that satisfies the installation specifications.
Note: Channel priority listed on the command decreases from left to right. So if you were to install base R r-base using the command below.
$ conda create --name rproject-env --channel defaults --channel conda-forge r-base
The first channel defaults would have a higher priority than the second conda-forge. This is true even, if the version number of the package is higher in the second channel.
Note: The bioconda team suggests that the conda-forge channel be a higher priority than the bioconda channel.
A Python package isn’t available on any Conda channel! What should I do?
If a Python package that you need isn’t available on any Conda channel, then you can use the default Python package manager Pip to install this package from PyPI. However, there are a few potential issues that you should be aware of when using Pip to install Python packages when using Conda.
First, Pip is sometimes installed by default on operating systems where it is used to
manage any Python packages needed by your OS. You do not want to use this pip to
install Python packages when using Conda environments.
(base) $ conda deactivate
$ which python
/usr/bin/python
$ which pip # sometimes installed as pip3
/usr/bin/pip
Second, Pip is also included in the Miniconda installer where it is used to install and
manage OS specific Python packages required to setup your base Conda environment. You
do not want to use this pip to install Python packages when using Conda environments.
$ conda activate
(base) $ which python
~/miniconda3/bin/python
$ which pip
~/miniconda3/bin/pip
Another reason to avoid installing packages into your
baseConda environmentIf your
baseConda environment becomes cluttered with a mix of Pip and Conda installed packages it may no longer function. Creating separate conda environments allows you to delete and recreate environments readily so you dont have to worry about risking your core Conda functionality when mixing packages installed with Conda and Pip.
If you find yourself needing to install a Python package that is only available via Pip, then
you should first install pip into your Conda environment and then use that pip to install
the desired package. Using the pip installed in your Conda environment to install Python packages
not available via Conda channels will help you avoid difficult to debug issues that frequently
arise when using Python packages installed via a pip that was not installed inside you Conda
environment.
Conda (+Pip): Conda wherever possible; Pip only when necessary
When using Conda to manage environments for your Python project it is a good idea to install packages available via both Conda and Pip using Conda; however there will always be cases where a package is only available via Pip in which case you will need to use Pip. Many of the common pitfalls of using Conda and Pip together can be avoided by adopting the following practices.
- Always explicitly install
pipin every Python-based Conda environment.- Always be sure your desired environment is active before installing anything using
pip.- Prefer
python -m pip installoverpip install; never usepipwith the--userargument.
Installing packages into Conda environments using
pippandas pandas is a fast, powerful, flexible and easy to use open source data analysis and manipulation too
Activate the conda environment ` python3-env
and usepipto installpandas`.Solution
The following commands will activate the ` python3-env
and install the python data analysis librarypandas`.$ conda activate python3-env ( python3-env) $ conda install pip ( python3-env) $ python -m pip install pandasFor more details on using
pipsee the official documentation.
Key Points
A package is a tarball containing system-level libraries, Python or other modules, executable programs and other components, and associated metadata.
A Conda channel is a URL to a directory containing a Conda package(s).
You can specific a conda channel using the option
--channelor add it to your.condarcIf a python package isn’t available on a conda channel you can install it into your environment using the python package installer
pip.
Sharing Environments
Overview
Teaching: 30 min
Exercises: 15 minQuestions
Why should I share my Conda environment with others?
How do I share my Conda environment with others?
How do I create an environment file that can be read by Windows, Mac OS, or Linux.
How do I specifying the package version in a Conda environment file.
Objectives
Understand why you would create an Conda environment file.
Create a Conda environment file in a text editor, specifying the channel, packages and their version.
Use the
conda envsubcommand to export a given environment to a environment file.
Reproducible research
Conda environments are useful when making bioinformatics projects reproducible. Full reproducibility requires the ability to recreate the system that was originally used to generate the results. This can, to a large extent, be accomplished by using a Conda environment file to make an environment with specific versions of the packages that are needed in the project. This environment file can then be shared with others users to reproduce your analysis environment containing software with the same version number.
Creating an environment file
Conda uses YAML (“YAML Ain’t Markup Language”) for writing its environment files. YAML is a human-readable language that is commonly used for configuration files and that that uses Python-style indentation to indicate nesting.
Creating your project’s Conda environment from a single environment file is a Conda “best practice”. Not only do you have a file to share with collaborators but you also have a file that can be placed under version control which further enhancing the reproducibility of your research project and workflow.
Default
environment.ymlfileNote that by convention Conda environment files are called
environment.yml. As such if you use theconda env createsub-command without passing the--fileoption, thencondawill expect to find a file calledenvironment.ymlin the current working directory and will throw an error if a file with that name can not be found.
Let’s take a look at an example environment.yml file to give you an idea of how to write your own environment files.
name: rnaseq-env
channels:
- conda-forge
- bioconda
- defaults
dependencies:
- salmon
- fastqc
- multiqc
The first line specifies a default name rnaseq-env for the environment, however this can be overidden on the command line. The second line specifies a list of channels, listed in priority order, that packages may need to be installed from. Finally the dependencies lists the most current and mutually compatible versions of the listed packages (including all required dependencies) to download.
The newly created environment would be installed inside the conda environment directory e.g. ~/miniconda3/envs/ directory, unless we specified a different path using conda create command line option --prefix or -p.
Since explicit versions numbers for all packages should be preferred a better environment file would be the following.
name: rnaseq-env
channels:
- conda-forge
- bioconda
dependencies:
- salmon=1.5
- fastqc=0.11
- multiqc=1.11
Note that we are only specifying the major and minor version numbers and not the patch or build numbers. Defining the version number by fixing only the major and minor version numbers while allowing the patch version number to vary allows us to use our environment file to update our environment to get any bug fixes whilst still maintaining significant consistency of our Conda environment across updates.
Always version control your
environment.ymlfiles!While you should never version control the contents of your
env/environment sub-directory, you should always version control yourenvironment.ymlfiles. Version controlling yourenvironment.ymlfiles together with your project’s source code means that you always know which versions of which packages were used to generate your results at any particular point in time.
Let’s suppose that you want to use the environment.yml file defined above to create a Conda environment in a sub-directory a project directory. Here is how you would accomplish this task.
$ cd ~/
$ mkdir rnaseq-project-2
$ cd rnaseq-project-2
Once your project folder is created, create an environment.yml file using your favourite editor for instance nano.
name: rnaseq-env
channels:
- conda-forge
- bioconda
dependencies:
- salmon=1.5
- fastqc=0.11
- multiqc=1.11
Finally create a new conda environment:
$ conda env create --prefix ./env --file environment.yml
$ conda activate ./env
Note that the above sequence of commands assumes that the environment.yml file is stored within your ` rnaseq-project-2` directory.
Automatically generate an environment.yml
We can automatically generate the contents of an environment file using the conda env export command. To export the packages installed into the previously created rnaseq-env you can run the following command:
$ conda env export --name basic-rnaseq-env
When you run this command, you will see the resulting YAML formatted representation of your Conda environment streamed to the terminal. Recall that we only listed three packages when we originally created basic-rnaseq-env yet from the output of the conda env export command we see that these packages result in an environment with a large number of dependencies!
To export this list into an environment.yml file, you can use --file option to directly save the resulting YAML environment into a file.
$ conda env export --name basic-rnaseq-env --file environment.yml
Make sure you do not have any other environment.yml file from before in the same directory when running the above command.
This exported environment file will however not consistently produce environments that are reproducible across Mac OS, Windows, and Linux. The reason is, that it may include operating system specific low-level packages which cannot be used by other operating systems.
If you need an environment file that can produce environments that are reproducible across Mac OS, Windows, and Linux, then you are better off just including those packages into the environment file that your have specifically installed using the --from-history option.
$ conda env export --name basic-rnaseq-env --from-history --file environment.yml
In short: to make sure others can reproduce your environment independent of the operating system they use, make sure to add the --from-history argument to the conda env export command.
Pip and conda env export
--from-historyPython packages installed via pip are not exported using the conda env export
--from-historyargument. You can add them to the environment YAML file using the keywordpip:followed by a list of python packages, For example;name: rnaseq-env channels: - conda-forge - bioconda dependencies: - salmon=1.5 - fastqc=0.11 - multiqc=1.11 pip: - pandas
Create a new environment from a YAML file.
Create a new project directory rnaseq-project-3 and then create a new
environment.ymlfile inside your project directory with the following contents.name: rnaseq-project3-env channels: - conda-forge - bioconda dependencies: - salmon=1.5 - fastqc=0.11 - multiqc=1.11Now use this file to create a new Conda environment. Where is this new environment created? Using the same
environment.ymlfile create a Conda environment as a sub-directory calledenv/inside a newly created project directory. Compare the contents of the two environments.Solution
To create a new environment from a YAML file use the
conda env createsub-command as follows.$ cd ~/ $ mkdir rnaseq-project-3 $ cd rnaseq-project-3 $ nano environment.yml $ conda env create --file environment.ymlThe above sequence of commands will create a new Conda environment inside the
envs_dirsdirectory. In order to create the Conda environment inside a sub-directory of the project directory you need to pass the--prefixto theconda env createcommand as follows.$ conda env create --file environment.yml --prefix ./envYou can now run the
conda env listcommand and see that these two environments have been created in different locations but contain the same packages.
Updating an environment
You are unlikely to know ahead of time which packages (and version numbers!) you will need to use for your research project. For example it may be the case that
- one of your core dependencies just released a new version (dependency version number update).
- you need an additional package for data analysis (add a new dependency).
- you have found a better visualization package and no longer need to old visualization package (add new dependency and remove old dependency).
If any of these occurs during the course of your research project, all you need to do is update the contents of your environment.yml file accordingly and then run the following command.
$ cd ~/
$ cd rnaseq-project-2
$ conda env update --prefix ./env --file environment.yml --prune
Note that the --prune option tells conda to remove any installed packages not defined in environment.yml
Rebuilding a Conda environment from scratch
When working with
environment.ymlfiles it is often just as easy to rebuild the Conda environment from scratch whenever you need to add or remove dependencies. To rebuild a Conda environment from scratch you can pass the--forceoption to theconda env createcommand which will remove any existing environment directory before rebuilding it using the provided environment file.$ conda env create --prefix ./env --file environment.yml --force
Update environment from environment.yml
Update the environment file from the previous exercise, rnaseq-project-3, by adding the package kallisto=0.46 and removing the salmon package. Then rebuild the environment.
Solution
The
environment.ymlfile should now look as follows.name: rnaseq-env channels: - conda-forge - bioconda dependencies: - fastqc=0.11 - multiqc=1.11 - kallisto=0.46You could use the following command, that will rebuild the environment from scratch with the new dependencies:
$ cd ~/rnaseq-project-3 $ conda env create --prefix ./env --file environment.yml --forceOr, if you just want to update the environment in-place with the new kallisto dependencies, you can use:
$ conda env update --prefix ./env --file environment.yml --prune
Restoring an environment
Conda keeps a history of all the changes made to your environment, so you can easily “roll back” to a previous version. To list the history of each change to the current environment:
$ conda activate basic-rnaseq-env
$ conda list --revisions
To restore environment to a previous revision:
$ conda install --revision=REVNUM or conda install --rev REVNUM.
For example,
$ conda install --revision=1
List revisions.
Activate the environment inside the
rnaseq-project-3and list the revisionsSolution
To create a new environment from a YAML file use the
conda env createsub-command as follows.$ cd ~/ $ cd rnaseq-project-3 $ conda activate ./env $ conda list --revisions
Key Points
Sharing Conda environments with other researchers facilitates the reproducibility of your research.
Conda environment files ,
environment.yml, describes your project’s software environment.
Configuring Conda
Overview
Teaching: 20 min
Exercises: 5 minQuestions
How can I configure conda ?
How can I see conda’s configuration values?
How can I modify conda’s configuration settings?
Objectives
Use the conda
config --showto display all configuration values.Modify the
.condarcfile using theconda configsub command.locate and view the contents of the
.condarcfile.
Configuration
Conda has a number of configuration setting which control how it performs.
To display and control these setting we can use the conda config subcommand.
To display all configuration settings run the config --show subcommand :
$ conda config --show
As you can see conda supports a large number of configuration options.
To show a single setting add the setting name after the conda config --show command.
For example, to show the list of channels conda searches run:
$ conda config --show channels
channels:
- defaults
By default conda only searches the defaults channel, this is why we had to include conda-forge and bioconda channels via the command line option --channel in the previous episode.
To get more information about an individual conda setting and its’ possible values run conda config --describe <option>. For Example;
conda config --describe channels
# # channels (sequence: primitive)
# # aliases: channel
# # env var string delimiter: ','
# # The list of conda channels to include for relevant operations.
# #
# channels:
# - defaults
.condarc
A user’s conda setting are store in the runtime configuration configuration file, .condarc. This file allows users to configure various aspects of conda including:
-
Where conda looks for packages
channels. -
Where conda lists known environments
envs_dirs. -
Whether to update the Bash prompt with the currently activated environment name
env_prompt. -
What default packages or features to include in new environments
create_default_packages.
Like the environment file the .condarc configuration file follows a simple YAML syntax
The .condarc file is not included by default, but it is automatically created in your home directory the first time you run the conda config command.
Creating or modify .condarc
To create or modify a .condarc file, enter the conda config command and use the modifier options --add, --set, --append , --prepend or --remove followed by the configuration key and a value .
conda config <modifier> <KEY> <VALUE>
Adding a configuration value
To add conda-forge to the list of channels we can use the --add, --append or --prepend modifier option:
For example, if we want to add a channel to our list of channels in our configuration file rather than specific it on the command line every time we can can use the conda config --add option modifier.
$ conda config --add channels conda-forge
This would add the conda-forge channel to the top of the channel list.
$ conda config --show channels
We can use the conda config modifier --append to add conda-forge to the end of the channel list, giving it the lowest priority.
$ conda config --append channels conda-forge
Warning: 'conda-forge' already in 'channels' list, moving to the bottom
$ conda config --show channels
To move a channel to the highest priority use the conda config --prepend modifier.
$ conda config --prepend channels conda-forge
Warning: 'conda-forge' already in 'channels' list, moving to the top
$ conda config --show channels
Note: It is generally best to have conda-forge as the highest priority channel as this will usually have the most up-to-date packages.
Adding the channels bioconda and conda-forge to .condarc.
Add the
biocondaandconda-forgechannels to your .condarc file. Giveconda-forgethe highest priority.Solution
To add the
biocondaandconda-forgechannel to your .condarc file use the command.$ conda config --add channels bioconda $ conda config --add channels conda-forgeThe above sequence of commands will add the channels to your
.condarc. Use the command below to show the channel priority order.$ conda config --get channels--add channels 'defaults' # lowest priority --add channels 'bioconda' --add channels 'conda-forge' # highest priority
Setting configuration settings
If our configuration setting has a single boolean or string value we can use conda config --set to set it.
For example, In a previous episode we set the command line prompt setting for conda using env_prompt.
$ conda config --describe env_prompt
The env_prompt setting takes a value of either '{prefix}', '{name}', and
'{default_env}'.
# # env_prompt (str)
# # Template for prompt modification based on the active environment.
# # Currently supported template variables are '{prefix}', '{name}', and
# # '{default_env}'. '{prefix}' is the absolute path to the active
# # environment. '{name}' is the basename of the active environment
# # prefix. '{default_env}' holds the value of '{name}' if the active
# # environment is a conda named environment ('-n' flag), or otherwise
# # holds the value of '{prefix}'. Templating uses python's str.format()
# # method.
# #
# env_prompt: '({default_env}) '
To set the env_prompt to the default value '({default_env})' we can run:
$ conda config --set env_prompt '({default_env})'
To change it back to the just the environment name, we can run.
$ conda config --set env_prompt '({name})'
Note:: You need to deactivate then reactivate the environment for the changes in env prompt to take effect.
Set conda
channel_priorityUse the
conda config --describeto investigate the settingchannel_priority. Set the channel_priority so that packages in lower priority channels are not considered if a package with the same name appears in a higher priority channel. Why would you want to do change this setting?Solution
$ conda config --describe channel_priority $ conda config --set channel_priority strict $ conda config --show channel_priorityUsing Strict channel priority can dramatically speed up conda operations and also reduce package incompatibility problems. This will be the default as of conda 5.0.
Editing the .condarc file manually
You can also use a text editor such as nano to directly edit the .condarc.
To show the location and contents of your .condarc file you can use the conda config --show-sources command.
$ conda config --show-sources
Note: If the .condarc file is in the root environment, it will override any in the home directory
Locate and view the
.condarcLocate your
.condarcfile. Using your favourite text editor look at the.condarc.Solution
$ conda config --show-sources$ nano ~/.condarc
Getting help
As with all conda commands you can use the --help option to get help.
For example, for a complete list of conda config commands run
$ conda config --help
Or see the command reference.
Key Points
The
.condarcis an optional configuration file that stores custom conda setting.You can use the
conda configsubcommand to add, set or remove configuration setting in the.condarcfile.You can also edit the contents of the
.condarcfile directly using a text editor.